Welcome
to my
website!
Paula Ramos-Silva
Data Steward and Teaching Coordinator (she/her, PhD)
Leiden University Medical Center
Bioinformatician and molecular biologist with experience in customer support. I support researchers on clinical and biomedical projects, focusing on scientific data management and stewardship.
Download my resume .
Interests
- Omics data
- Bioinformatics
- Data management and stewardship
- Molecular Evolution
- Spatial Biology
- Education
Education
-
PhD in Biology - Bioinformatics, 2013
University of Amsterdam
-
MSc + BSc in Biological Engineering, 2007
Instituto Superior Tecnico
Professional experience
Data Steward and Teaching Coordinator
Technical Application Scientist
I support researchers in resolving complex issues, from sample prep to instrument troubleshooting and multi-omics data analyses.
Marie Skłodowska-Curie Researcher
I used omics approaches in planktonic calcifiers, to better understand the mechanisms governing their shell formation and impacts of ocean acidification on this process.
Postdoctoral Fellow in Computational Genomics
I used comparative genomics to trace the origin and macroevolution of sporulation in bacteria.
Projects
Funded research projects
.js-id-evospore
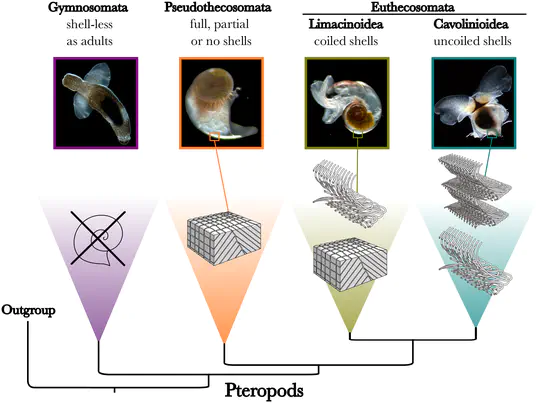
Evolution of Planktonic Gastropod Calcification
Ocean acidification due to increasing CO2 reduces the amount of carbonate ions in seawater, putting a wide range of marine calcifiers at risk. Sea butterflies and sea elephants are among the most vulnerable calcifiers since they inhabit the open ocean and make thin shells of calcium carbonate. Genetic tools for these planktonic snails are scarce making it difficult to predict their future in acidified waters. The Marie Skłodowska-Curie project EPIC aimed at studying the molecular processes by which shell formation evolved in planktonic gastropods over macroevolutionary scales, and to inform on their potential to adapt under long-term ocean acidification.

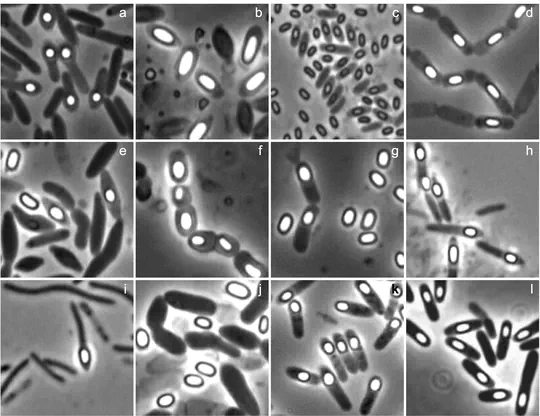
Understanding the evolutionary history and the diversity of bacterial endosporulation
The project EvoSpore was funded by the Portuguese Foundation for Science and Technology. The goal was to uncover the origin and evolution of bacterial endosporulation through comparative genomics. Endosporulation is a developmental pathway that initiates from the vegetative cell and culminates with the formation of highly resistant, dormant spores. Spores are everywhere, they allow environmental persistence, dissemination and, for pathogens, are infection vehicles. We found that for the model Bacillus subtilis and for the pathogen Clostridioides difficile, sporulation was shapped by two major evolutionary transitions. The first one was at the base of the Firmicutes and the second one at the base of the B. subtilis group and within the Peptostreptococcaceae family, which includes C. difficile. We also found that early and late sporulation regulons have been coevolving and that sporulation genes entail greater innovation in B. subtilis with many Bacilli lineage-restricted genes. In contrast, C. difficile more often recruits new sporulation genes by horizontal gene transfer, which reflects both its highly mobile genome, the complexity of the gut microbiota, and an adjustment of sporulation to the gut ecosystem.

Publications
All publications available on Google Scholar

